SemMed Database
The SemMed Database is a biomedical medical database deployed using the MySQL Database Management System.
This post is about generating a Knowledge Graph that manifests as a Semantic Web from this database by leveraging the multi-model database management system capabilities of Virtuoso.
Relational Data Details
ENTITY table
This table contains entity information whose data come from ENTITY output generated using full fielded output. It includes the following data fields:
ENTITY_ID— Auto-generated primary key for each unique entitySENTENCE_ID— The foreign key toSENTENCEtableCUI— The CUI of the entityNAME— The preferred name of the entityTYPE— The semantic type of the entityGENE_ID— The EntrezGene ID of the entityGENE_NAME— The EntrezGene name of the entityTEXT— The text in the utterance that maps to the entitySTART_INDEX— The first character position (in document) of the text denoting the entityEND_INDEX— The last character position (in document) of the text denoting the entitySCORE— The confidence score
PREDICATION table
Each record in this table identifies a unique predication. The data fields are as follows:
PREDICATION_ID— Auto-generated primary key for each unique predicationSENTENCE_ID— Foreign key to theSENTENCEtablePMID— The PubMed identifier of the citation to which the predication belongsPREDICATE— The string representation of each predicate (for exampleTREATS,PROCESS_OF)SUBJECT_CUI— The CUI of the subject of the predicationSUBJECT_NAME— The preferred name of the subject of the predicationSUBJECT_SEMTYPE— The semantic type of the subject of the predicationSUBJECT_NOVELTY— The novelty of the subject of the predicationOBJECT_CUI— The CUI of the object of the predicationOBJECT_NAME— The preferred name of the object of the predicationOBJECT_SEMTYPE— The semantic type of the object of the predicationOBJECT_NOVELTY— The novelty of the object of the predication
SENTENCE table
This table contains information about individual sentences from PubMed citations and includes the following data fields:
SENTENCE_ID— Auto-generated primary key for each sentencePMID— The PubMed identifier of the citation to which the sentence belongsTYPE—'ti'for the title of the citation,'ab'for the abstractNUMBER— The location of the sentence within the title or abstractSENTENCE— The actual string or text of the sentenceSECTION_HEADER— Section header name of structured abstract (from Version 3.1)NORMALIZED_SECTION_HEADER— Normalized section header name of structured abstract (from Version 3.1)
Knowledge Graph Creation Steps
-
Download the SemMed Database MySQL data dump file (see data sources table)
-
Load the MySQL database dump file into a MySQL database
-
Attach MySQL Database to Virtuoso via ODBC
-
Generate RDF Views using Attached Tables.
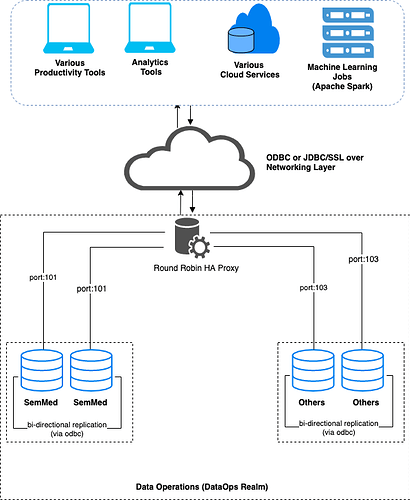
The Virtuoso installation architecture for achieving this task could be as depicted below.
At this stage, you have basic RDF 3-Tuple based Relations (RDF Graphs) generated from N-Tuple Relations (SQL Tables) in place that reflect the following:
- Each Table is treated as a Class (Entity Type)
- Each Table Record is treated as an Instance (or Individual) of the Class for that Table
- Each Column Name is a Relation Name (RDF sentence Predicate)
- Each Column Value is a Relation Value (RDF sentence Object)
- Each Column is also associated with a Class via an
rdfs:domainRelation - Each Column Value is associate with a Class via an
rdfs:rangeRelation
Based on the above, you can generate a domain-specific Knowledge Graph to, for example, associate genes with diseases.
-
Generate of additional triples using SPARQL
INSERT -
Run test queries to verify the state and quality of the resulting Knowledge Graph
Example Queries
Genes – Live Link
PREFIX semmed-gene: <http://semmed.demo.openlinksw.com/schemas/Gene#>
SELECT DISTINCT *
FROM <urn:semmed:rdf:data:new3>
FROM <urn:semmed:rdf:data:genes3>
WHERE
{
?s a semmed-gene:this ;
owl:sameAs ?o
}
LIMIT 100
Genes associated with Asthma – [Live Link]
PREFIX associated_with: <http://semmed.demo.openlinksw.com/terms/associated_with#>
PREFIX copd: <http://semmed.demo.openlinksw.com/about/c0024117#>
PREFIX asthma: <http://semmed.demo.openlinksw.com/about/c0004096#>
PREFIX semmed-gene: <http://semmed.demo.openlinksw.com/schemas/Gene#>
SELECT DISTINCT *
FROM <urn:semmed:rdf:data:genes3>
FROM <urn:semmed:rdf:data:new3>
WHERE
{
?s a semmed-gene:this ;
rdfs:label ?genName ;
associated_with:this asthma:this .
}
Genes associated with Asthma, but not COPD 1 – [Live Link]
PREFIX associated_with: <http://semmed.demo.openlinksw.com/terms/associated_with#>
PREFIX copd: <http://semmed.demo.openlinksw.com/about/c0024117#>
PREFIX asthma: <http://semmed.demo.openlinksw.com/about/c0004096#>
PREFIX semmed: <http://semmed.demo.openlinksw.com/schemas/semmed/>
PREFIX semmed-gene: <http://semmed.demo.openlinksw.com/schemas/Gene#>
SELECT DISTINCT *
FROM <urn:semmed:rdf:data:genes3>
FROM <urn:semmed:rdf:data:new3>
WHERE
{
?s a semmed-gene:this ;
rdfs:label ?genName ;
associated_with:this asthma:this .
FILTER ( NOT EXISTS { ?s associated_with:this copd:this }
)
}
Genes associated with Asthma, but not COPD 2 (using GROUP_CONCAT) – [Live Link]
PREFIX associated_with: <http://semmed.demo.openlinksw.com/terms/associated_with#>
PREFIX copd: <http://semmed.demo.openlinksw.com/about/c0024117#>
PREFIX asthma: <http://semmed.demo.openlinksw.com/about/c0004096#>
PREFIX semmed: <http://semmed.demo.openlinksw.com/schemas/semmed/>
PREFIX semmed-gene: <http://semmed.demo.openlinksw.com/schemas/Gene#>
SELECT DISTINCT ?s
( GROUP_CONCAT ( ?geneName , "\n" ) AS ?name )
FROM <urn:semmed:rdf:data:genes3>
FROM <urn:semmed:rdf:data:new3>
WHERE
{
?s a semmed-gene:this ;
rdfs:label ?geneName ;
associated_with:this asthma:this .
FILTER ( NOT EXISTS { ?s associated_with:this copd:this }
)
}
Genes associated with Asthma and COPD – [Live Link]
PREFIX associated_with: <http://semmed.demo.openlinksw.com/terms/associated_with#>
PREFIX copd: <http://semmed.demo.openlinksw.com/about/c0024117#>
PREFIX asthma: <http://semmed.demo.openlinksw.com/about/c0004096#>
PREFIX semmed: <http://semmed.demo.openlinksw.com/schemas/semmed/>
PREFIX semmed-gene: <http://semmed.demo.openlinksw.com/schemas/Gene#>
SELECT DISTINCT *
FROM <urn:semmed:rdf:data:genes3>
FROM <urn:semmed:rdf:data:new3>
WHERE
{
?s a semmed-gene:this ;
rdfs:label ?genName ;
associated_with:this asthma:this ,
copd:this .
}
Entity Reconciliation Demo Sequence Example
Using B3GNT2 as the industry standard identifier of interest.
Here’s LOD, as a superior resolver:
-
B3GNT2Text Search results filtered by type which reveals Bio2RDF effect (this Data Space was the original resolver for Biological Identifiers)