Uniprot Knowledge Graph & ChatGPT Fine-tuning using SPARQL
Q: What is Uniprot, and why is its SPARQL Query Service important?
A:
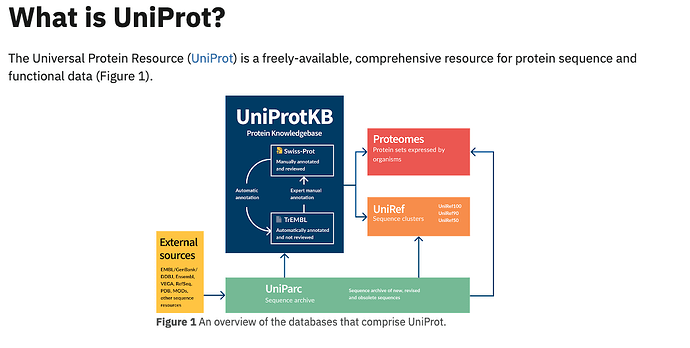
Uniprot is a comprehensive resource for protein sequence and annotation data². It consists of three databases: the UniProt Knowledgebase (UniProtKB), which contains manually reviewed and automatically annotated protein entries; the UniProt Reference Clusters (UniRef), which clusters protein sequences at different levels of identity to reduce redundancy; and the UniProt Archive (UniParc), which stores all publicly available protein sequences².
The Uniprot SPARQL Query Service is a web service that allows users to query and access Uniprot data using the SPARQL query language¹. SPARQL is a standard language for querying and manipulating RDF data, which is a format that represents data as a graph of entities and relationships¹. The Uniprot SPARQL Query Service supports the SPARQL 1.1 Standard and provides a web interface where users can enter and execute queries, as well as download the results in various formats².
The Uniprot SPARQL Query Service is important because it enables users to perform complex and customized queries on Uniprot data that are not possible with the web interface or the API. For example, users can query and combine data from different Uniprot databases, such as UniProtKB, UniRef, UniParc, and taxonomy, as well as from other SPARQL endpoints hosted by the EBI, the SIB, or other sources¹. This provides a valuable low-cost alternative to building your own data warehouse. The Uniprot SPARQL Query Service also provides examples of simple and advanced queries that users can modify and reuse for their own purposes².
Source: Conversation with Bing, 6/2/2023
(1) UniProt. https://sparql.uniprot.org/.
(2) SPARQL for UniProt. https://legacy.uniprot.org/help/sparql.
(3) UniProt. https://www.uniprot.org/.
(4) SPARQL | UniProt - EMBL-EBI. SPARQL | UniProt.
In this post, I will demonstrate the powerful benefits that arise from fine-tuning the GPT4 Large Language Model (the LLM) used by ChatGPT (set to GPT 4.0 mode) using SPARQL query templates derived from the publicly available Uniprot sample queries collection.
Large Language Models & Natural Language Processing
A lot has been written about this topic, and from my own experimentation the utility of LLMs boils down to their ability deliver pretty revolutionary natural language processing due to a profound understanding of sentence structure, rules of grammar, and “parts of speech” semantics.
Here’s a simple demonstration, using ChatGPT, that provides easy to understand clarity to the statement above,
Q: How many variations of the following sentence can you generate?
Find all Natural Variant Annotations if associated via an evidence tag to an article with a pubmed identifier.
A:
Sure, here are a few variations on your sentence:
- Locate all Natural Variant Annotations tied to an article by an evidence tag, if the article has a PubMed identifier.
- If an article possesses a PubMed identifier, obtain all Natural Variant Annotations connected to it through an evidence tag.
- Ascertain all Natural Variant Annotations related to an article via an evidence tag, given that the article holds a PubMed identifier.
- Discover all Natural Variant Annotations linked by an evidence tag to any article carrying a PubMed identifier.
- Uncover all Natural Variant Annotations related to an article via an evidence tag, provided the article features a PubMed identifier.
- Retrieve all Natural Variant Annotations that are associated through an evidence tag to an article, if it is identified by PubMed.
- Identify all Natural Variant Annotations associated with an article, through an evidence tag, if the article is referenced by a PubMed identifier.
- Look for all Natural Variant Annotations associated with an article via an evidence tag, if the article has a PubMed ID.
- Search for all Natural Variant Annotations linked to an article via an evidence tag, assuming the article comes with a PubMed identifier.
- Get all Natural Variant Annotations connected to an article using an evidence tag, if the article has a PubMed identifier.
Linked Data Principles, Knowledge Graphs, and a Semantic Web
Linked Data Principles simply imply the use of HTTP-based hyperlinks to unambiguously name entities, entity types, and entity relationship types. These entities, types, and relationships provide the framework for constructing digital sentences that describe anything of interest using RDF (Resource Description Framework).
This approach to structured data representation enables the construction of knowledge graphs that manifest a semantic web rife with connections that facilitate explicit and/or serendipitous discovery. Basically, a variant of the Web on steroids.
Combined Force Implications
This implies the ability to map a myriad of natural language sentences to a single, structured query expressed in SPARQL that targets the high-quality, publicly-accessible knowledge graph provided by UniProt. UniProt’s knowledge graph comprises more than 100 billion entity relationships, which are deployed using Linked Data principles. The knowledge graph is hosted on a single Virtuoso DBMS instance.
How?
Let’s resume our interactions with ChatGPT to demonstrate the effects of SPARQL Query based fine-tuning that ultimately ups the ante regarding response quality.
- Go to the Uniprot SPARQL examples page
- Take an example query of interest
- Use the query description and actual query text to create a simple {Prompt};{Response} structured fine-tuning template
- Register template with your ChatGPT session
- Construct prompts that are similar to what was used in your template definition – which is where the sentence similarity comprehension magic of the GPT4 LLM kicks in
Query Description Example
Find all Natural Variant Annotations if associated via an evidence tag to an article with a pubmed identifier
Associated SPARQL Query Text Example
SELECT
?accession
?annotation_acc
?pubmed
WHERE
{
?protein a up:Protein ;
up:annotation ?annotation .
?annotation a up:Natural_Variant_Annotation .
?linkToEvidence rdf:object ?annotation ;
up:attribution ?attribution .
?attribution up:source ?source .
?source a up:Journal_Citation .
BIND(SUBSTR(STR(?protein),33) AS ?accession)
BIND(IF(CONTAINS(STR(?annotation), "#SIP"), SUBSTR(STR(?annotation),33), SUBSTR(STR(?annotation),36))AS?annotation_acc)
BIND(SUBSTR(STR(?source),35) AS ?pubmed)
}
Fine-tuning Template Example
Structure: {Query-Description-Text};{Query}
Find all Natural Variant Annotations if associated via an evidence tag to an article with a pubmed identifier;
SELECT
?accession
?annotation_acc
?pubmed
WHERE {
?protein a up:Protein ;
up:annotation ?annotation .
?annotation a up:Natural_Variant_Annotation .
?linkToEvidence rdf:object ?annotation ;
up:attribution ?attribution .
?attribution up:source ?source .
?source a up:Journal_Citation .
BIND(SUBSTR(STR(?protein),33) AS ?accession)
BIND(IF(CONTAINS(STR(?annotation), "#SIP"), SUBSTR(STR(?annotation),33), SUBSTR(STR(?annotation),36))AS?annotation_acc)
BIND(SUBSTR(STR(?source),35) AS ?pubmed)
}
Here’s a session transcript snippet demonstrating the fine-tuning effects of the example template.
Q: Given the template.
Find all Natural Variant Annotations if associated via an evidence tag to an article with a pubmed identifier;
SELECT
?accession
?annotation_acc
?pubmed
WHERE {
?protein a up:Protein ;
up:annotation ?annotation .
?annotation a up:Natural_Variant_Annotation .
?linkToEvidence rdf:object ?annotation ;
up:attribution ?attribution .
?attribution up:source ?source .
?source a up:Journal_Citation .
BIND(SUBSTR(STR(?protein),33) AS ?accession)
BIND(IF(CONTAINS(STR(?annotation), "#SIP"), SUBSTR(STR(?annotation),33), SUBSTR(STR(?annotation),36))AS?annotation_acc)
BIND(SUBSTR(STR(?source),35) AS ?pubmed)
}
Search for all Natural Variant Annotations linked to an article via an evidence tag, assuming the article comes with a PubMed identifier, using the uniprot sparql endpoint
A:
Given your SPARQL query and the template, here’s the variation you’re looking for:
“Query the uniprot SPARQL endpoint to find all Natural Variant Annotations that are linked to an article through an evidence tag, assuming that the article is associated with a PubMed identifier.”
In a technical environment, the sentence might be written as:
“Using the uniprot SPARQL endpoint, execute a SELECT query to fetch all Natural Variant Annotations tied to an article via an evidence tag, given the article is attached with a PubMed identifier.”
And here’s the SPARQL query in a format ready for use:
PREFIX up: <http://purl.uniprot.org/core/>
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
SELECT
?accession
?annotation_acc
?pubmed
WHERE {
?protein a up:Protein ;
up:annotation ?annotation .
?annotation a up:Natural_Variant_Annotation .
?linkToEvidence rdf:object ?annotation ;
up:attribution ?attribution .
?attribution up:source ?source .
?source a up:Journal_Citation .
BIND(SUBSTR(STR(?protein),33) AS ?accession)
BIND(IF(CONTAINS(STR(?annotation), "#SIP"), SUBSTR(STR(?annotation),33), SUBSTR(STR(?annotation),36)) AS ?annotation_acc)
BIND(SUBSTR(STR(?source),35) AS ?pubmed)
}
Q: Redo, but returning a hyperlink that denotes the generated SPARQL query .
A:
Certainly. Here is a revised version:
Utilize the uniprot SPARQL endpoint to query all Natural Variant Annotations associated with an article via an evidence tag, provided that the article is equipped with a PubMed identifier. Access the generated SPARQL query here.
Q:
Redo, in response to the question:
If an article possesses a PubMed identifier, obtain all Natural Variant Annotations connected to it through an evidence tag.
A:
Sure. Here is a revised version:
If an article possesses a PubMed identifier, it’s possible to retrieve all Natural Variant Annotations associated with it through an evidence tag. This can be done by querying the uniprot SPARQL endpoint. You can access the generated SPARQL query here.
Conclusion
It is common knowledge that ChatGPT (and other LLM-based Smart Agents) have response accuracy challenges when dealing with the kind of fine-grained knowledge held in knowledge graphs such as Uniprot. However, it is not as widely known that SPARQL Query based fine-tuning can provide a powerful, yet loosely coupled, solution to this challenge. This post has demonstrated, using a simple example, the use of SPARQL Query based fine-tuning to improve the response accuracy of ChatGPT.
Important Takeaway
Anyone can build on this effort by simply creating more query templates from the Uniprot examples collection, or by repeating the process across 200+ publicly accessible SPARQL Query Services endpoints that make up the massive LOD Cloud Knowledge Graph.